※ User Guide:
Frequently Asked Questions:
1. Q: How to read the GPS-PBS results?
A: Here we use the human protein RAF-1 as the example. After clicking "Submit", the prediction results of 14-3-3 binding sites with high threshold are shown as follows:
<1>. The table of the GPS-PBS results (Page 1)
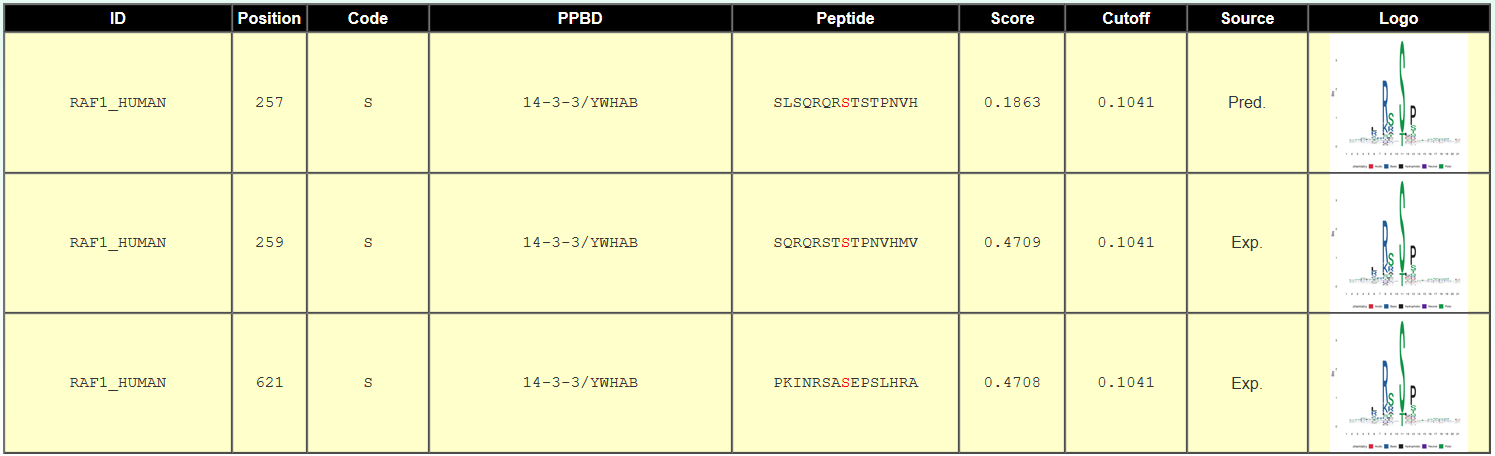
ID: The name/id of the protein sequence that you input to predict.
Position: The position of the site which is predicted to be bound.
Code: The residue which is bound by PPBDs.
PPBDs: The cognate PPBDs which is predicted to bind the site.
Peptide: The predicted phosphopeptide with 7 amino acids upstream and 7 amino acids downstream around the modified residue.
Score: The value calculated by GPS-PBS to evaluate the potential of binding.
Cutoff: The cutoff value under the threshold. Different threshold means different precision, sensitivity and specificity.
Source: Whether the interactions is validated by experiment, "Exp." means YES, while "Pred." means NO.
Logo: The sequence logo of this phosphopeptide.
2. Q: Is GPS-PBS accurate?
A: Yes, but not all. Systematically matching these sites to the phosphoprotein-binding domains (PPBDs) that bind to them is an extremely hard task. If the training data is more enough, the prediction is more satisfying and accurate. But for many PPBDs, the training data set are constrained which made the performance lower. However, in comparison, GPS-PBS achieved a higher accuracy than other existing tools.
3. Q: How to choose the cut-off values and the thresholds?
A: Firstly, we calculated the theoretically maximal false positive rate (FPR) for each PPBDs cluster. The three thresholds of GPS-PBS were decided based on calculated FPRs.For phosphoserine/phosphothreonine (pS|pT) group, the high, medium and low thresholds were established with FPRs of 2%, 6% and 10%. And for phosphotyrosine (pY), the high, medium and low thresholds were selected with FPRs of 4%, 9% and 15%. in substrates.
4. Q: What's the meaning of False Positive Rate (FPR)?
A: To estimate the false positive rate (FPR) of prediction, we randomly retrieved 10,000 PBP(10, 10) peptides from eukaryotic proteomes to construct a near-negative data set. However, precise calculation of FPR is unavailable due to lack of a "gold-standard" negative data set. The process was repeated twenty times and the average FPR was calculated by GPS-PBS as the theoretically maximal FPR.
5. Q: I have a few questions which are not listed above, how can I contact the authors of GPS-PBS?
A: Please contact the major author: Dr. Yu Xue for details.



